摘要
盐度是影响湖泊细菌群落最重要的环境因子之一,揭示盐度对细菌群落变化的影响对于深入理解水生生态系统细菌群落具有重要意义。本研究以青藏高原西南部的22个湖泊为对象,探讨33个沉积物样本的细菌丰富度、均匀度和群落组成,分析其随盐度梯度的变化规律,阐明其潜在的环境驱动因素。结果表明,变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)和放线菌门(Actinobacteria)是青藏高原湖泊沉积物中的优势细菌门类,相对丰度分别为46.54%、12.41%和7.23%,总和达到整个细菌门类的66.18%。对于细菌α多样性,物种丰富度和均匀度均随盐度增加呈显著下降趋势;对于细菌β多样性,基于非度量多维排列分析,根据盐度可以将细菌群落分布分为3个显著的独立簇,分别对应淡水、咸水和高盐环境。Mantel检验结果表明,相较于地理距离,湖泊沉积物细菌群落Bray-Curtis不相似性随盐度的变化更为显著。方差分解分析和随机森林分析结果表明,细菌物种丰富度、均匀度和群落组成变化的最佳预测因子为盐度和磷酸盐等化学因素,整体贡献率分别为72.98%、59.82%和60.83%,其中盐度是丰富度、均匀度和群落组成变化的主要驱动因素,其相对贡献率分别为45.47%、79.18%和79.50%。基于结构方程模型进一步分析表明,湖泊沉积物细菌丰富度、均匀度和群落组成变化均受到盐度直接或间接的影响。本研究结果充分探讨了盐度对细菌群落变化的影响,揭示了青藏高原湖泊沉积物细菌组成及其环境驱动因素,可为理解湖泊生态系统细菌群落对环境变化的响应提供科学支撑。
Abstract
Salinity is one of the most important environmental factors affecting bacterial communities in lake sediments. Revealing the impact of salinity on bacterial community changes is of great significance for a deeper understanding of bacterial communities in aquatic ecosystems. This study focused on 22 lakes in the southwest of the Qinghai-Tibet Plateau, explored the bacterial richness, evenness and community composition of 33 sediment samples, analyzed their changes with salinity, and elucidates their potential environmental driving factors. The results showed that Proteobacteria, Bacteroidetes and Actinobacteria were the dominant bacterial phyla in the sediment of lakes, with relative abundances of 46.54%, 12.41% and 7.23%, respectively, totaling 66.18% of the entire bacterial phyla. For bacterial α diversity, species richness, and evenness showed a significant downward trend with increasing salinity. For bacterial β diversity, based on non-metric multidimensional permutation analysis, bacterial community distribution could be divided into three significant independent clusters by salinity, corresponding to freshwater, saline and high salinity environments, respectively. The Mantel test results indicated that compared to geographical distance, the Bray-Curtis dissimilarity of lake sediment bacterial communities varied more significantly with salinity. The results of variance decomposition analysis and random forest analysis indicated that the best predictive factors for changes in bacterial species richness, evenness and community composition were chemical factors such as salinity and phosphate, with overall contribution rates of 72.98%, 59.82% and 60.83%, respectively. Among them, salinity was the main driving factor for changes in richness, evenness and community composition, with relative contribution rates of 45.47%, 79.18% and 79.50%, respectively. Furthermore, relying on structural equation modeling, changes in bacterial richness, evenness and community composition in lake sediment were directly or indirectly affected by salinity. The results of this study fully explored the impact of salinity on bacterial community changes and revealed the bacterial composition and environmental driving factors of sediment in lakes on the Qinghai-Tibet Plateau. This provided scientific support for understanding the response of bacterial communities in lake ecosystems to environmental changes.
阐明微生物多样性、群落组成及其潜在驱动因素是生态学研究的关键之一[1]。细菌是水生生态系统的重要组成部分,是湖泊中有机碳、硫、含氮化合物和金属转化的主要贡献者,在生态系统食物网和养分循环中发挥着不可替代的作用[2-3]。生物多样性,包括α多样性和β多样性,涉及多个组成部分,如物种丰富度、均匀度和群落结构组成等[4]。物种丰富度主要用于衡量一个生态系统微生物物种的数量,而均匀度则衡量生态系统中不同物种之间数量的差异度[5-6]。研究表明,物种丰富度和均匀度通常呈正相关关系,但是由于环境因素的影响,丰富度和均匀度也存在负向关系[7]。β多样性,即物种组成的时空变化,对于理解群落构建的扩散限制及其环境响应机制具有重要意义[8]。例如,研究发现群落相似性随环境距离和地理距离的增加而显著衰减[9],并表明β多样性随盐度的变化可能受到环境异质性、人类活动和气候变化的影响[10]。
盐度被认为是影响湖泊[11]、沿海水域[12]、河口[13]和海洋[14]等细菌群落的最重要环境因素之一。与大型生物相比,微生物如细菌对盐度的响应更为复杂和敏感[15]。一方面,高盐度可以通过提高渗透压,降低细胞代谢酶活性,进而破坏微生物酶的结构并抑制微生物生长[16];另一方面,湖泊沉积物细菌敏感地响应于盐度变化,继而产生生理适应机制以抵消盐度的不利影响[17]。此外,盐度极大地影响水体细菌的群落组成[18-19]。例如,γ-变形菌纲和β-变形菌纲主要在淡水样本中占主导地位[20],α-变形菌纲主要在海洋样本中占主导地位[21],并且α-变形菌纲和γ-变形菌纲的比例随着盐度的变化保持不变[22]。因此,详细探讨盐度对细菌多样性和群落组成变化的影响,对维持湖泊生态系统功能和稳定性具有重要意义。
青藏高原是世界海拔最高、中国最大的高原,被称为“世界屋脊”“第三极”,气候为干旱或半干旱气候,年平均降水量小于400 mm,年平均气温低于5℃,拥有世界上数量最多的高海拔湖泊(平均海拔超过4000 m)[23]。湖泊约32843个,总面积约为43151.08 km2[24-25]。青藏高原独特的地理位置和气候特征使湖泊细菌群落面临多种环境变化的挑战,如盐度、海拔高度、pH值、气温和降水等[26-27],尤其是青藏高原湖泊的盐度变化范围极为广泛(0.1~400 g/L)[28]。因此,青藏高原为研究盐度对高原湖泊沉积物细菌群落的影响提供了理想的环境条件。然而,对于青藏高原湖泊沉积物细菌的物种多样性和群落组成如何随盐度变化以及是否受其他环境因子的影响,相关研究还较少。
本研究收集来自青藏高原西南部22个湖泊的33个样本,基于测序和生物信息学分析手段获得青藏高原湖泊沉积物细菌群落的α多样性和β多样性,探讨青藏高原湖泊沉积物细菌丰富度、均匀度和群落组成随盐度的变化,进一步阐明其潜在的环境驱动因素。具体而言,本文旨在回答以下问题:青藏高原湖泊的细菌群落组成如何随盐度梯度变化?影响湖泊沉积物细菌丰富度、均匀度和群落组成的主要环境变量还有哪些?它们的相对贡献分别是多少?盐度如何直接或间接影响湖泊沉积物细菌丰富度、均匀度和群落组成?
1 材料与方法
1.1 研究区域和采样
本研究以青藏高原西南部22个湖泊为对象,共采集了33个沉积物样品。其中,在班戈错(XT09)、班公湖(XT10)、阿木错(XT11)、佩枯错(XT12)、郭扎错(XT13)、才多茶卡(XT25)、巴松错(XT27)、然乌湖(XT28)、令戈错(XT29)、美玛错(XT30)、错鄂(XT31)和伊布茶卡(XT32)各设置1个采样点,在松木希错(XT03、04)、玛旁雍错(XT05、06)、邦达错(XT07、08)、阿鲁错(XT15、16)、泉水湖(XT20、21)、普尔错(XT22、23)、鄂雅错琼(XT01、24)、拉昂错(XT14、26)和龙木错(XT02、33)各设置2个采样点,在结则茶卡(XT17~19)设置3个采样点(图1a,附表Ⅰ)。所选湖泊覆盖范围为28°58′12″~35°0′36″N,79°48′36″~96°49′48″E。2021年8月,通过采水器从湖泊表层(顶部50 cm)收集1 L表层水样品并带回实验室,用于后续进行水样的理化指标分析,并现场测量水深和透明度。用柱状采样器采集表层沉积物样本,并取柱状沉积物样本的最顶部1 cm于封装袋,将每个表层沉积物样品分成两个子样品,分别于-20℃和4℃下储存用于实验室分析。前者用于提取DNA和微生物群落分析,后者用于理化性质测定。
1.2 理化指标测定
水体样品通过0.45 μm的滤膜过滤后,利用连续流动分析仪(Skalar SA1000)测定水体氨氮(NH3-N)、硝态氮(NO-3-N)、亚硝态氮(NO-2-N)和磷酸盐(PO3-4-P)。按照标准方法测定水体总氮(TN)和总磷(TP)[29]。用多参数水质测定仪(YSI,6000,USA)测定水样的温度、pH值、溶解氧浓度、盐度和电导率。对于沉积物样品,将表层沉积物样品冷冻干燥至恒重,研磨成细粉,并通过100目筛(孔径为0.150 mm)进行总碳(TC)、TN分析[30]。沉积物TC和TN含量通过元素分析仪(Flash EA 1112,Italy)进行测量。用便携式全球定位系统记录地理空间参数,包括经度、纬度和海拔。所有检测到的环境参数都测定3次,取平均值用于后续分析。
1.3 气候数据提取
从国家青藏高原科学数据中心的《青藏高原气候空间数据集》(https://doi.org/10.11888/AtmosphericPhysics.tpe.49.file)下载青藏高原地区的气候数据。根据样本的地理位置经纬度提取2020年所研究湖泊的年平均气温(MAT,℃)和年平均降水量(MAP,mm)。在ArcGIS(v10.2)软件中利用ArcToolbox中的Spatial Analyst Tools工具完成提取。
1.4 细菌群落DNA提取和测序
根据制造商的说明,使用MoBio PowerSoil DNA分离试剂盒(MoBio,Carlsbad,USA)从沉积物样品中提取细菌等微生物的DNA。通过聚合酶链式反应(PCR)用引物515F(5′-GTG YCA GCM GCC GCG GTA A-3′)和806R(5′-GGA C TA CNV GGG TWT CTA AT-3′)一式三份扩增16S rRNA基因的V4高变区,然后混合重复[7]。使用脚本“pick_open_reference_otus.py”将序列处理到微生物生态学软件(QIME2 v2022.8)中[31]。本文使用Denoiser算法对长度超过450 bp的序列进行去噪[32],接着使用基于种子的UCLUST算法以97%的相似性阈值将它们聚类为操作分类单元(OTU)[33]。
1.5 统计分析
首先,利用线性模型探究盐度与细菌门类丰度的关系[34]。使用“vegan”包计算物种丰富度(Richness)和均匀度(Evenness),以评估研究区细菌群落的α多样性[35]。使用非度量多维标度(NMDS)确定各采样点群落结构的显著变化。生物差异通常随着地理距离和两个个体群落之间的盐度差异而增加[36]。使用 Bray-Curtis 差异性研究细菌β 多样性如何随盐度变化,以量化成样点之间群落组成的差异,并且通过 Mantel 检验确定其显著性[37]。基于Bray-Curtis矩阵,探讨随盐度的增加细菌群落组成的差异,并使用“vegan” 包进行可视化。NMDS第1轴和第2轴上两个位点(即NMDS1和NMDS2)的距离代表群落组成的差异[38]。使用 NMDS1 来描述群落组成,表明物种数据中最重要的变化。
其次,利用相关性分析探究环境驱动因素与物种丰富度、均匀度和群落结构组成3个生物多样性指标之间的关系,分别以Richness、Evenness和NMDS1为指标。在下面的分析中,选择性地排除了具有高相关性(Spearman's r>0.7)的变量,以使共线性最小化,确保结果更具可解释性。基于距离的冗余分析(redundancy analysis,RDA)将具有高方差膨胀因子 (VIF>20) 的变量排除在RDA之外[39]。同时,进行Mantel检验(999次排列)以确定群落与环境矩阵之间的关系显著性。Vegan 包中的“envfit”函数用于从 RDA 图中排除对群落结构没有重大影响的因素[39]。
随后,方差分解分析(variance partitioning analysis,VPA)被应用于研究层次变量对多样性、均匀性和群落组成的独立或组合贡献[40]。将实测环境因子分为3类:物理因素(包括水温、水深、透明度、经度和纬度)、化学因素(包括盐度、溶解氧、电导率、pH值、TN、TP、NH3-N、NO-3-N、NO-2-N和PO3-4-P)以及气候因素(包括年均气温和年均降水量)。为了进一步量化环境因素对3个预测群落指标的影响,使用 “randomForest” 包,应用随机森林模型。随机森林是一种机器学习算法,它通过创建一组具有二进制除法的分类树来扩展标准分类和回归树方法[41]。“randomForest”函数用于执行回归。通过平均森林中 2000 棵树的预测精度下降来计算每个预测变量的重要性。当预测变量的数据随机排列时,通过计算观测值与预测值之间的均方误差的增加来确定每个预测变量的减少[39]。
最后,使用结构方程模型(SEM)检验盐度、其他环境因子、营养盐、地理位置和气候因素对湖泊沉积物细菌丰富度、均匀度和群落组成间的直接或间接影响。根据现有数据结果构建先验模型,使用“lavaan”包,进行CFA分析,对信效度、模型拟合指数进行相关分析,选择影响沉积物细菌群落变化的主要理化性质。常用的模型拟合评价指标包括P值、比较拟合指数(CFI)、均方根误差(RMR)和标准化均方根误差(SRMR)[42-43]。良好的拟合标准为0.05<P≤1.00、0.95≤CFI≤1、0≤RMR≤0.08和0≤SRMR≤0.05[42,44]。
上述各项统计方法于R软件(4.2.3版)中分析(http://cran.r-project.org)。
2 结果与分析
2.1 环境理化性质
青藏高原湖泊覆盖范围广(图1a),具有明显的盐度变化,研究区湖泊的盐度范围为0.04~97.18 g/L,为研究盐度对高原湖泊沉积物细菌物种丰富度、均匀度和群落组成的影响提供了理想的环境条件。其中,研究区共9个湖泊14个采样点的水体为淡水(盐度<1 g/L);8个湖泊10个采样点的水体为咸水(1 g/L <盐度<35 g/L);5个湖泊9个采样点的水体为高盐水(盐度> 35 g/L)。采样湖泊的pH值为3.70~10.12,水温为7.40~16.80℃,海拔为3434~5092 m,年平均气温为-10.68~2.61℃,年均降水量为43.81~810.31 mm。水体透明度范围为0.25~8.50 m,平均值为3.37 m;电导率范围为56.80~95287.00 μS/cm,平均值为26924.64 μS/cm;DO浓度范围为3.23~7.09 mg/L,平均值为5.39 mg/L;TN浓度范围为0.29~10.18 mg/L,平均值为2.01 mg/L;TP浓度范围为0.01~0.70 mg/L,平均值为0.07 mg/L;NH3-N浓度范围为0.01~5.58 mg/L,平均值为0.45 mg/L;NO-3-N浓度范围为0.01~1.39 mg/L,平均值为0.22 mg/L;NO-2-N浓度范围为0~0.05 mg/L,平均值为0.008 mg/L;PO3-4-P浓度范围为0~0.64 mg/L,平均值为0.06 mg/L(附表Ⅰ)。
线性回归分析结果表明,沿着纬度梯度,部分环境理化性质呈现显著的上升或者下降趋势。具体而言,随着纬度的增加,盐度(R2=0.31,P<0.01,图1b)、海拔(R2=0.53,P<0.001,图1c)和NO-2-N(R2=0.28,P<0.05,图1h)呈显著上升趋势,而年平均气温(R2=0.60,P<0.001,图1d)、年均降水量(R2=0.65,P<0.001,图1e)、水深(R2=0.17,P<0.05,图1f)、水温(R2=0.14,P<0.05,图1g)和pH值(R2=0.25,P<0.05,图1i)则呈显著下降趋势。
2.2 细菌群落组成
通过16S rRNA测序分析,对每个OTU中最丰富的序列进行分类标注,并获得各采样点OTU的分类信息。在青藏高原湖泊33个采样点的沉积物样品中,共检测出76个门类、195个纲类的95532个OTUs。各采样点沉积物中最丰富的前10个细菌种群在门水平上的分布和相对物种丰度如图2a所示。变形菌门(Proteobacteria,OTU数为43463)、拟杆菌门(Bacteroidetes,OTU数为8229)和放线菌门(Actinobacteria,OTU数为6552)的相对丰度均值分别为46.54%、12.41%和7.23%,总和超过65%,是青藏高原湖泊的主要优势细菌门。其中,变形菌门相对丰度最高,在纲水平上主要包括γ-变形菌纲(Gamma-proteobacteria,53.83%,OTU数为23400)、δ-变形菌纲(Delta-proteobacteria,28.31%,OTU数为12307)和α-变形菌纲(Alpha-proteobacteria,16.17%,OTU数为7028);在科水平上主要包括鱼立克次体科(Piscirickettsiaceae,5.1%,OTU数为2223)、脱硫杆菌科(Desulfobacteraceae,5.0%,OTU数为2186)和红杆菌科(Rhodobacteraceae,4.1%,OTU数为1796)。其次是拟杆菌门,在纲水平上主要包括拟杆菌纲(Bacteroidia,51.04%,OTU数为3344)、腐败螺旋菌纲(Saprospiria,14.48%,OTU数为949)、噬纤维菌纲(Cytophagia,10.03%,OTU数为657)和黄杆菌纲(Flavobacteriia,9.58%,OTU数为628);在科水平上主要包括腐败螺旋菌科(Saprospiraceae,8.3%,OTU数为549)、噬纤维菌科(Cytophagaceae,7.6%,OTU数为501)和噬几丁质菌科(Chitinophagaceae,5.6%,OTU数为373)。放线菌门相对丰度也较高,在纲水平上主要包括酸微菌纲(Acidimicrobiia,40.86%,OTU数为2596)和嗜热油菌纲(Thermoleophilia,25.49%,OTU数为1620);在科水平上主要包括盖勒氏菌科(Gaiellaceae,13.6%,OTU数为865)、腈基降解菌科(Nitriliruptoraceae,5.7%,OTU数为366)和Iamiaceae(2.1%,OTU数为135)。
图1青藏高原湖泊采样点分布及其环境因子
Fig.1Distribution of sampling sites and environmental factors in lakes on the Qinghai-Tibet Plateau
对于细菌群落,除变形菌门、拟杆菌门、厚壁菌门(Firmicutes)、热变形菌门(Thermoproteota)和螺旋体门(Spirochaetes)外,其他优势门的相对丰度随盐度升高而呈显著下降趋势(P<0.01,图2b~k)。其中,放线菌门、绿弯菌门(Chloroflexi)、酸杆菌门(Acidobacteria)和疣微菌门(Verrucomicrobia)均呈一次线性下降趋势,且绿弯菌门下降幅度最大(Slope=-0.76),放线菌门下降幅度最小(Slope=-0.45),而浮霉菌门(Planctomycetes)的相对丰度随着盐度梯度呈先上升再下降的趋势,总体表现为显著的驼峰状趋势(P<0.01)。此外,拟杆菌门和热变形菌门表现出U型趋势(P<0.05)。
2.3 细菌群落多样性随盐度梯度的变化
通过计算湖泊沉积物细菌物种丰富度和均匀度,将它们与盐度进行线性回归分析,探讨细菌群落α多样性随盐度增加的变化。结果表明,细菌物种丰富度(R2=0.52,P<0.001,图3a)和均匀度(R2=0.81,P<0.001,图3b)都随着盐度的增加呈现显著下降趋势。
此外,基于NMDS分析探讨湖泊沉积物细菌群落组成的不相似性。将采样湖泊通过盐度划分为3组,分别为淡水湖泊(FL,盐度<1g/L)、咸水湖泊(SL,1 g/L ≤ 盐度≤35 g/L)和高盐湖泊(HSL,盐度> 35 g/L)。研究结果表明,根据盐度将细菌群落分布形成了3个显著的独立簇(Stress=0.084,图4a),分别对应3种盐度的湖泊环境。Mantel检验结果进一步表明,Bray-Curtis不相似性随着盐度和地理距离的增加而增加(盐度:Mantel R2=0.42,P<0.001;地理距离:Mantel R2=0.12,P<0.05),表明湖泊沉积物细菌群落组成在所研究的湖泊中显示为盐度/地理距离—衰变关系。相较于地理距离,湖泊沉积物细菌群落Bray-Curtis差异与盐度更加密切相关(图4b、c)。

图2各样点在门分类学水平上基于相对丰度的细菌群落组成及其随盐度的变化 (XT01、XT02、XT24、XT28和XT30 5个样品未获得测序结果)
Fig.2The composition of bacterial communities at various taxonomic levels based on relative abundance and their variation with salinity

图3细菌物种丰富度和均匀度与盐度的线性回归关系
Fig.3Linear regression relationship of bacterial communities between species richness (or evenness) and salinity
2.4 物种丰富度、均匀度和群落组成的驱动因素
为了深入了解湖泊沉积物细菌物种丰富度、均匀度和群落组成的驱动因素,共考虑了3个维度的环境驱动因素,包括物理因素(如海拔、水深、透明度和水温等)、化学因素(盐度、电导率、pH值、总有机碳、TN、TP、营养盐和溶解氧等)和气候因素(年均气温和年均降水量等)。
首先,通过RDA探讨了影响青藏高原湖泊沉积物细菌群落组成变化的主要环境因素(图4d)。结果表明,前两个主成分 RDA1 和 RDA2 分别解释了细菌群落组成变化的 47.47% 和 20.59%。物理、化学和气候因素在解释青藏高原湖泊沉积物细菌群落组成变化中具有重要作用。其中,盐度(P<0.001)、PO3-4-P(P<0.05)、透明度(P<0.05)以及年均气温(P<0.05)等环境因素对整个群落组成都表现出显著影响。
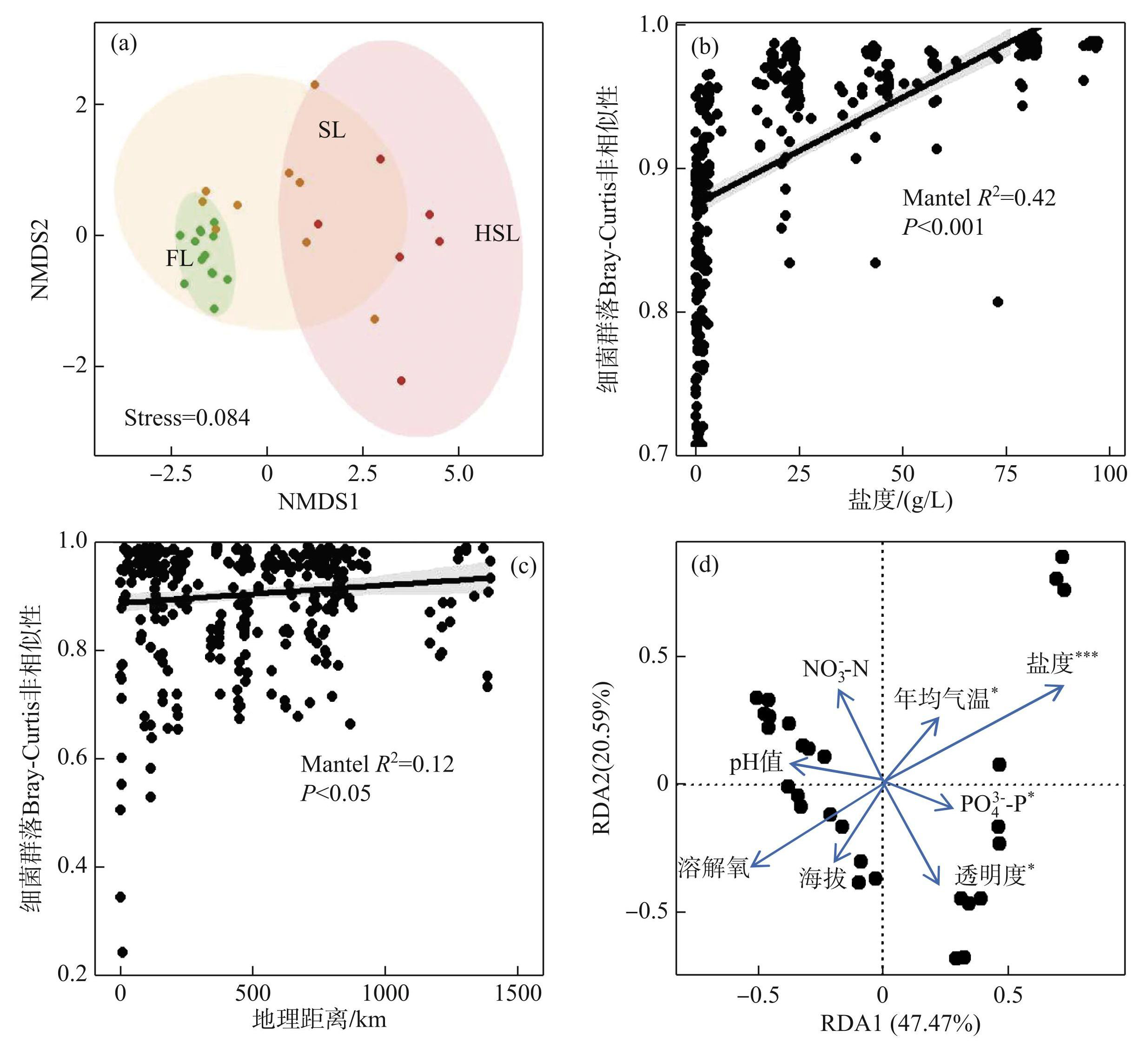
图4细菌群落与环境因子的NMDS分析、与盐度和地理距离的Bray-Curtis 差异性分析以及冗余分析(FL:淡水湖泊;SL:咸水湖泊;HSL:高盐湖泊)
Fig.4NMDS analysis, Bray-Curtis difference analysis with salinity and geographic distance and redundancy analysis of bacterial communities and environmental factors (FL: freshwater lake; SL: salinity lake; HSL: high salinity lake)
其次,分析了细菌群落多样性指标(包括Richness、Evenness和NMDS1)与各物理因素、化学因素和气候驱动因素的Spearman相关性(图5)。结果表明,化学因素中的盐度(Spearman's r>0.15,P<0.01)、电导率(Spearman's r>0.12,P<0.01)、溶解氧(Spearman's r>0.18,P<0.01)、TN(Spearman's r>0.12,P<0.01)和NO-2-N(Spearman's r>0.11,P<0.01)都是显著影响Richness、Evenness和NMDS1的关键因素。其中,与物种丰富度和均匀度相比,群落结构组成与环境因子的相关性更强。具体而言,盐度(r=0.62)、电导率(r=0.58)、TN(r=0.56)、NO-2-N(r=0.47)和 PO3-4-P (r=0.42)均更加显著地影响细菌群落结构(P<0.05);而物种丰富度(r=0.52)和均匀度(r=0.48)主要受溶解氧的影响(P<0.01)。
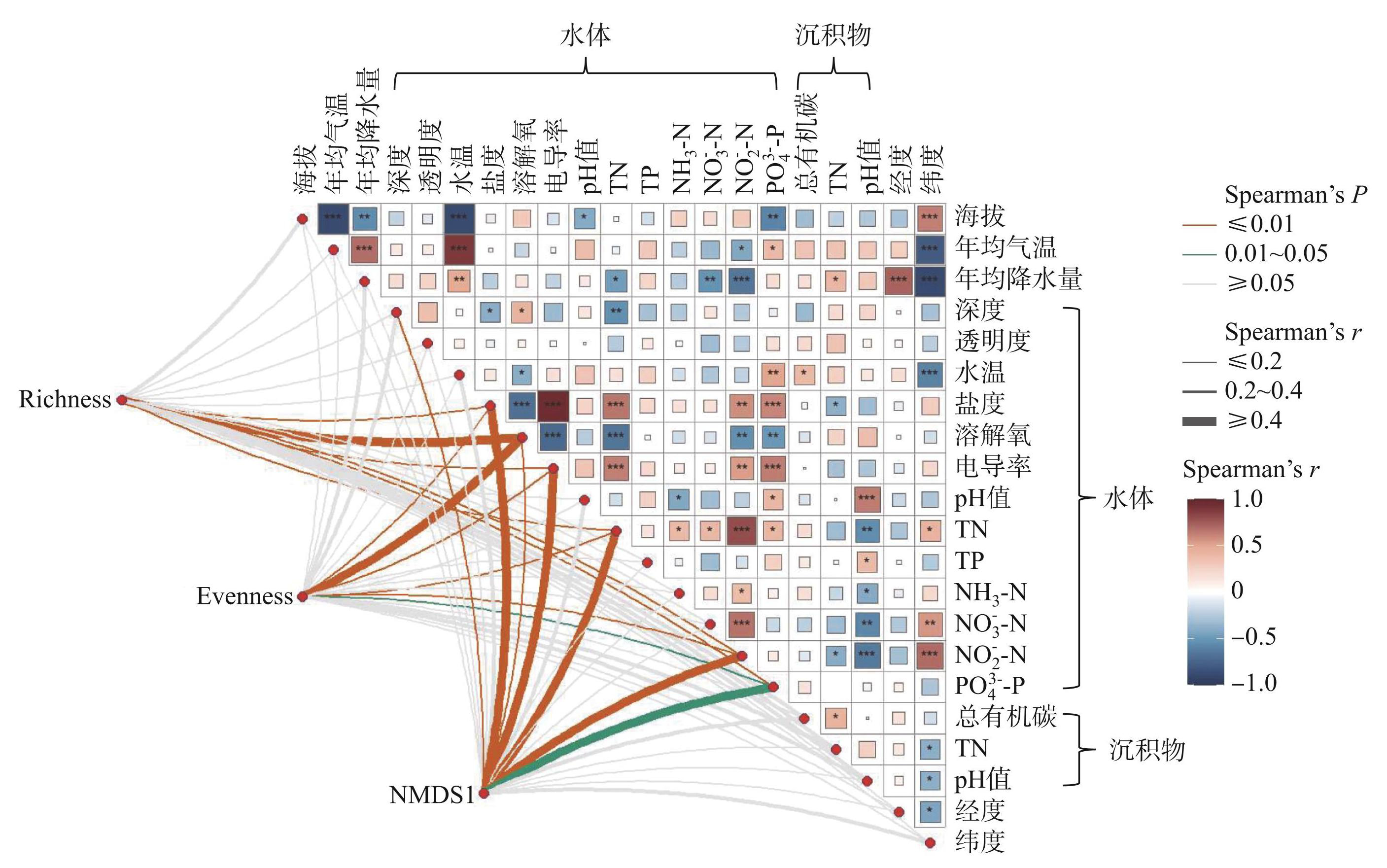
图5环境因子与细菌物种丰富度、均匀度和群落组成的Spearman相关性关系
Fig.5Spearman correlation between environmental factors and bacterial species richness, evenness and community composition
随后,使用方差膨胀因子(VIF)来筛选环境自变量之间共线性程度较低的因素,并通过方差分解和随机森林分析(图6)探讨了筛选出的物理因素、化学因素和气候因素中对细菌物种丰富度、均匀度和群落组成变化的最佳预测因子以及不同环境因素的相对贡献。结果表明,化学因素是影响青藏高原湖泊沉积物细菌物种丰富度、均匀度和群落组成变化的最佳预测因子,其相对总解释量分别为72.98%、59.82%和60.83%(图6a~c)。其中,PO3-4-P和盐度是细菌物种丰富度变化的主要驱动因素,相对贡献率分别为52.16%和45.47%(图6d);盐度和PO3-4-P对细菌物种均匀度变化的影响最大,相对贡献率分别为79.18%和11.83%(图6e)。此外,盐度和PO3-4-P对群落组成变化的影响也最大,相对贡献率分别为79.50%和14.90%(图6f),这与图4d的RDA分析结果一致。
最后,通过构建SEM来分析盐度、其他环境因子、营养盐、地理位置和气候对湖泊沉积物细菌多样性和群落组成间的直接和间接影响(图7)。SEM结果表明,盐度是控制湖泊沉积物细菌群落组成的最重要因素,营养盐对沉积物细菌群落组成有较强影响(图7a~c)。具体而言,影响沉积物细菌丰富度的关键驱动因素是盐度(标准化效应(SE)=-0.63),一方面盐度直接影响细菌丰富度(路径系数β=-0.94,P<0.001),另一方面盐度通过影响溶解氧浓度而间接影响细菌丰富度(β=-0.92,P<0.001)。营养盐也是影响细菌丰富度的重要驱动因素,包括PO3-4-P(β=-0.21,P<0.05)、NO-2-N(β=-0.62,P<0.01)和NH3-N(β=0.55,P<0.001)。影响沉积物细菌均匀度的关键驱动因素是盐度(SE=-0.47)和NO-2-N(SE=-0.62)。总有机碳(β=0.44,P<0.01)和pH(β=-0.40,P<0.01)也是影响细菌均匀度的重要驱动因素。影响沉积物细菌群落组成的关键驱动因素是盐度(SE=-0.57)和NO-3-N(SE=-0.49)。盐度一方面直接影响沉积物细菌群落组成(β=-0.48,P<0.01),另一方面通过影响TN而间接影响细菌群落组成(β=0.72,P<0.05)。此外,NO-2-N(β=0.42,P<0.01)和纬度(β=0.41,P<0.01)也是影响细菌群落组成的重要驱动因素。
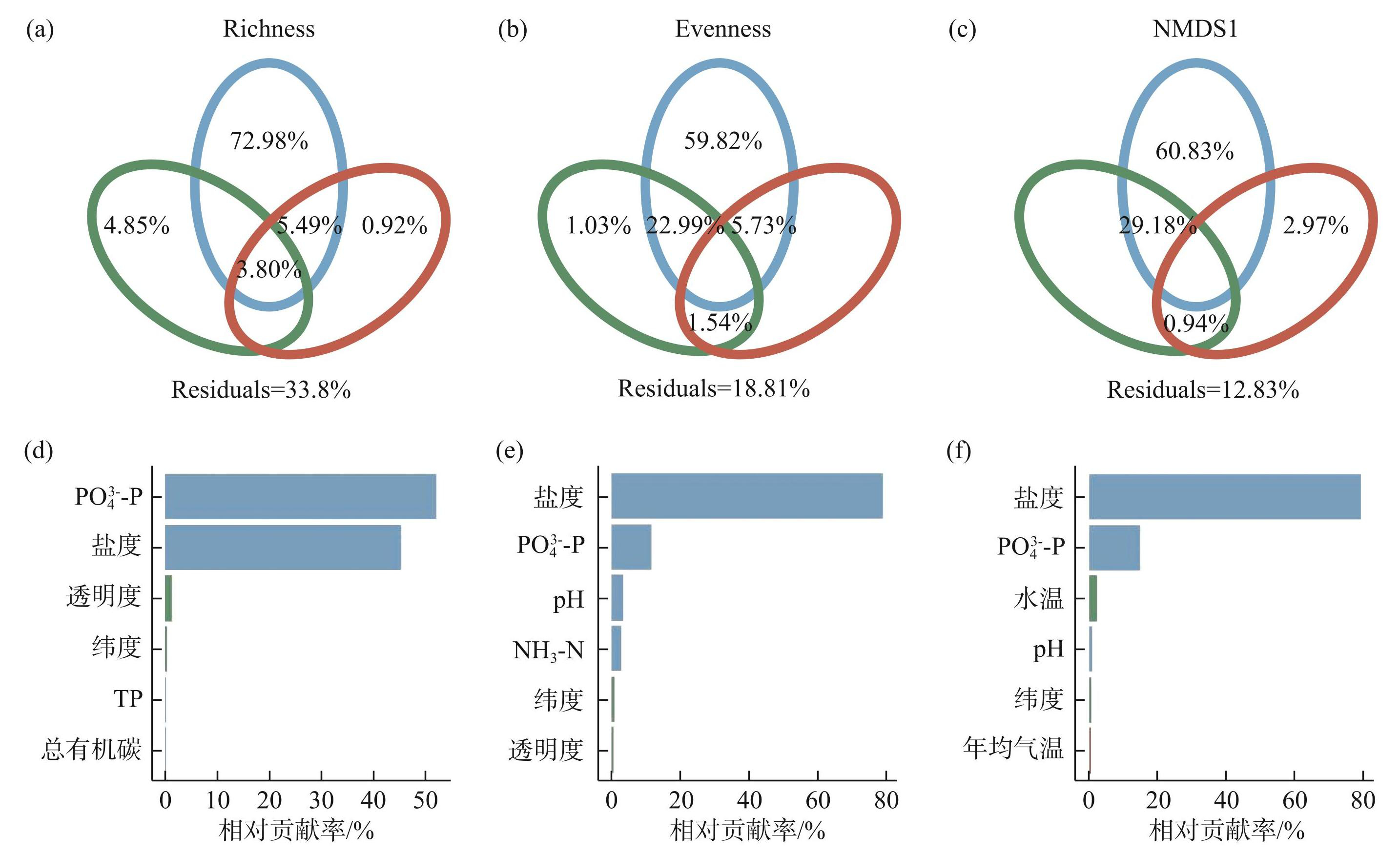
图6细菌物种丰富度、均匀度和群落组成与环境因子的VPA分析和随机森林分析 (图中绿色表示物理因素,蓝色表示化学因素,红色表示气候因素)
Fig.6VPA analysis and random forest analysis of bacterial species richness, evenness, and community composition with environmental factors (In figures, green represents physical factors, blue represents chemical factors, and red represents climate factors)
3 讨论
尽管湖泊生态系统中的细菌物种多样性和群落组成越来越受到关注,但是在大尺度上细菌群落随盐度的变化以及环境驱动因素对细菌群落影响的相对重要性研究仍然较少。本研究收集了来自青藏高原西南部22个湖泊的33个样本,基于测序研究了青藏高原西南部湖泊沉积物细菌群落的α多样性和β多样性,包括物种丰富度、均匀度和群落组成,并探讨了其盐度格局以及潜在环境驱动因素。结果表明:(1)变形菌门、拟杆菌门和放线菌门是青藏高原湖泊沉积物细菌的主要优势门类。(2)高原湖泊沉积物细菌的物种丰富度和均匀度均随盐度的升高而降低。(3)高原湖泊沉积物细菌的物种丰富度、均匀度和群落组成主要受化学因素影响,如盐度和营养盐。(4)湖泊沉积物细菌丰富度、均匀度和群落组成变化均受到盐度直接或间接的影响。
3.1 细菌群落组成特征
数据显示,研究区域中细菌群落的主要优势菌门是变形菌门(46.54%),其次是拟杆菌门(12.41%)和放线菌门(7.23%)。变形菌门在整个研究区域中相对丰度较高,且随盐度变化呈非显著趋势(图2a),这说明变形菌可能耐受或适应较广泛的环境梯度,包括温度(从寒冷到炎热)、pH值(从酸性到碱性)和盐度(从淡水、咸水到高盐水)等[45-47]。比如在高温环境下,变形菌具有丰富的活性肽酶,不仅用于控制和调节蛋白质质量和转换内肽酶,还可以用于裂解细胞壁和降解细胞破裂后周围环境中的有机物,因此它们可以积极地适应高温环境[48]。此外,研究表明在水生生态系统(比如湖泊)中细菌群落通常以变形菌门为主,其次是拟杆菌门、厚壁菌门、放线菌门和蓝细菌门[49-50],本研究结果与其一致。在湖泊中变形菌门主要包括γ-变形菌纲(53.83%)、δ-变形菌纲(28.31%)和α-变形菌纲(16.17%),这些分类群通常在盐水或其他高盐环境中占主导地位[51]。
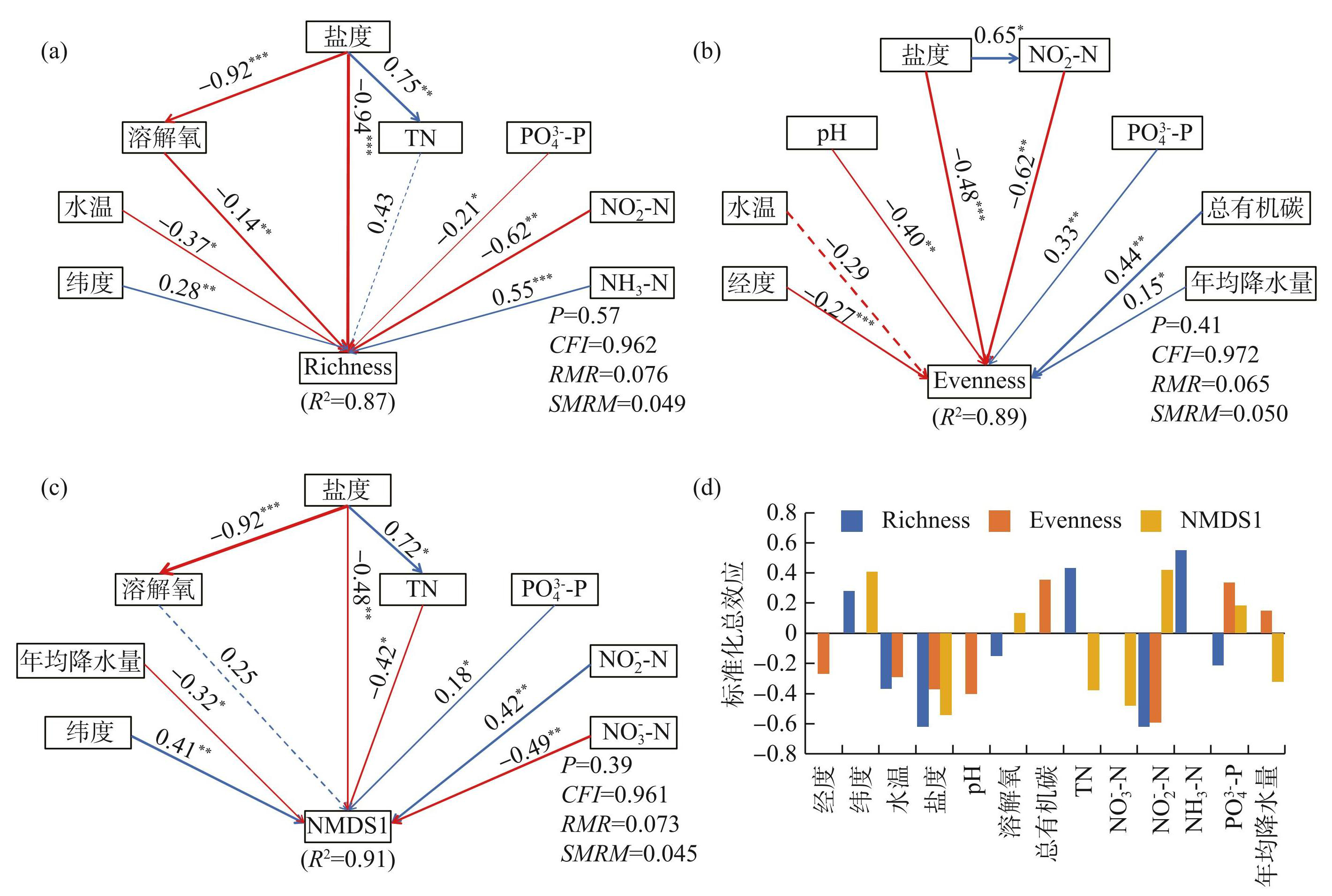
图7环境因素对湖泊沉积物细菌丰富度(a)、均匀度(b)和群落组成(c)直接或间接影响的结构方程模型(SEM)以及这些因素在SEM中的标准化总效应(d)(蓝色箭头表示正相关关系,红色箭头表示负相关关系;实线箭头表示影响显著,虚线箭头表示影响不显著;箭头宽度表示标准化路径系数的强度;R2表示模型解释的方差;*P<0.05,**P<0.01,***P<0.001)
Fig.7Structural equation modeling (SEM) of the direct or indirect effects of environmental factors on bacterial richness (a) , evenness (b) and community composition (c) in lake sediments, as well as the standardized total effects of these factors in SEM (d)
拟杆菌门是第二丰富的门类,它与湖泊沉积物中的营养物质转化有关[52]。结果发现,拟杆菌门在青藏高原湖泊中的相对丰度与盐度密切相关,并且随着盐度的升高而显著增加(图2c)。研究表明,在水生环境中拟杆菌与高盐度有关[53-55]。拟杆菌门中常见的嗜盐菌,如红盐杆菌(Salinibacter ruber),在盐度高达30%的结晶池中占原核生物群落的25%[54]。耐盐细菌在盐水中生长与生存常采取以下策略[56]:例如通过特定的膜或细胞壁结构最大限度地减少盐的吸收[57];将离子排出细胞从而调节体内的渗透压[58];产生适应高浓度溶质离子的蛋白质和酶[59]。此外,本研究结果还发现放线菌门也普遍存在于湖泊沉积物细菌群落中,并且随着盐度的升高而显著减少(图2d),这与Lew等的研究结果相同,即放线菌喜好淡水湖泊环境,通常在淡水生态系统的浮游细菌中占主导地位,并且放线菌数量随着盐度的增加而显著下降[15]。
3.2 物种丰富度、均匀度和群落结构的盐度格局
对于α多样性,本研究结果显示,湖泊沉积物细菌物种丰富度和均匀度均随盐度增加呈显著下降的趋势。从细菌门的水平考虑,主要优势门类(包括变形菌门、放线菌门、浮霉菌门、绿弯菌门、酸杆菌门和疣微菌门)的丰度都随着盐度的上升而下降(图2a)。盐度与细菌群落α多样性指数之间的负相关也可能与部分已有生态学原理一致,即预计更极端的环境微生物群落多样性更低[60-61]。这可能是由于盐度的增加影响了细胞的生理特性,增强其渗透潜力,降低了微生物的活性和生物量[62],从而显著影响细菌群落α多样性。而细菌物种均匀度的下降可能是因为高盐度的环境通过选择性地富集某些细菌谱系来影响细菌群落组成,同时排除具有低盐度耐受性的细菌谱系[61]。例如,有研究表明在变形菌门中,γ-变形菌纲(Gamma-proteobacteria)和α-变形菌纲(Alpha-proteobacteria)的相对丰度分别与盐度呈显著负相关和正相关[63-65]。
盐度对细菌多样性的影响可能还与尺度有关。与淡水湖相比,有些咸水湖表现出相似甚至更高的细菌多样性水平[66]。这在一定程度上表明小幅度增加盐度可能会使生态系统具有更大的群落多样性,而盐度增加过大则会显著降低群落多样性[67]。这可能归因于营养物质和资源的增加,或者是耐盐细菌和淡水细菌在中等盐度下可利用生态位的增加[68]。此外,在大多数封闭的青藏高原湖泊中,水的供应来自降水和冰川,水的流失由蒸发造成[69]。因此,水力停留时间是影响盐度的因素之一,这可能也是盐度影响细菌群落多样性的原因。
对于β多样性,Mantel检验表明,Bray-Curtis差异随着盐度和地理距离增加都呈显著增加趋势,表明细菌群落组成在青藏高原湖泊沉积物中显示出显著的盐度/地理距离—衰变关系。且相较于地理距离,湖泊沉积物细菌群落Bray-Curtis差异与盐度的关系更加密切(图4b、c)。这一结果与Yang等的研究一致,在该研究中,相比地理距离,盐度显著影响丰富和稀有的微生物群落结构[10]。然而,Santini等发现湖泊之间的地理距离是影响微生物群落组成变化的主要因素[70],这可能是由于空间因素对微生物分布格局的影响还与尺度有关。例如,有研究表明在小空间尺度(<500 km)上,环境变量是影响细菌群落的主要因素;而在大空间尺度(>1000 km)上,空间因子在微生物群落变化的形成中起重要作用[71]。
3.3 细菌多样性和群落组成的环境驱动因素
VPA分析和随机森林分析结果表明,化学因素(包括盐度和PO3-4-P等)是细菌物种丰富度(72.98%)、均匀度(59.82%)和群落组成(60.83%)变化的最佳预测因子(图6a~c)。因此,除了盐度,营养物质的可用性在湖泊沉积物细菌群落中也起着重要作用[58]。Shi等研究发现磷酸盐对细菌多样性有显著影响,黄杆菌(Flavobacterium)和脱硫单胞菌(Desulfuromonas)是参与磷元素等重要生物地球化学循环的细菌[72]。然而尽管无机氮(N)和磷(P)对细菌生长至关重要,但是它们的富集会降低土壤和水生生态系统中的细菌多样性[73],研究表明细菌多样性通常在营养物质的中等水平达到峰值[74]。
此外,SEM结果表明,湖泊沉积物细菌丰富度、均匀度和群落组成变化均受到盐度直接或间接的影响,同时还受到营养盐、地理位置和气候因素的直接影响(图7a~c)。盐度通过影响NO-2-N或TN而间接影响细菌均匀度或群落组成。研究表明,在青藏高原不同盐度的湖泊沉积物中,反硝化速率和产物具有差异,如高盐湖泊中反硝化速率较低,N2O为主要产物;随着盐度降低,反硝化速率增加,主要反硝化产物转变为N2[75]。氮被认为是细菌代谢过程的重要常量营养物质,包括光合作用、呼吸、能量储存和转移以及细胞分裂等过程[76]。部分研究表明,TN是影响沉积物细菌群落和微生物功能结构最重要的环境因子[77-78]。这可能是由于与氮相关的微生物,如反硝化菌和硝化菌,是湖泊沉积物细菌群落的优势物种,它们在湖泊生态系统中影响着微生物群落的组成和分布[79]。盐度还通过影响溶解氧而间接影响细菌群落。较高的盐度会降低水体的溶解氧浓度,这可能是因为盐的存在会增加水分子之间的相互作用力,使水分子变得更紧密,从而减小溶解氧的空间[80]。而地理位置对细菌群落的直接影响主要表现在两个方面。一是地理距离的变化导致水的理化因子(如盐度和营养盐)发生变化[81];二是地理距离的增加导致生境间地理隔离,阻碍了生境间细菌的扩散[82],进而导致湖泊间细菌群落的差异。此外,气候因素(如年均气温和年均降水量)也是影响湖泊沉积物细菌群落的重要因子。已有研究表明,在广泛空间尺度上,气候因素是形成微生物多样性的重要驱动因素[83]。较高的年均气温、年均降水量都可能导致径流将更多的陆地微生物输送到湖泊中[84],从而影响这些湖泊中细菌群落组成结构的空间变异性。
4 结论
本研究以青藏高原西南部22个湖泊为对象,分析了33个表层沉积物样本的细菌群落,通过线性回归分析、NMDS分析、RDA分析、VPA分析、随机森林分析和SEM构建,综合探讨了盐度对青藏高原湖泊沉积物细菌丰富度、均匀度和群落组成的直接或间接影响以及其他潜在的环境驱动因素,主要得出如下结论:
1)变形菌门、拟杆菌门和放线菌门是青藏高原湖泊沉积物中的优势细菌门类,相对丰度分别为46.54%、12.41%和7.23%。变形菌门在整个研究区域中相对丰度较高,且随盐度变化呈非显著趋势,这说明变形菌可能耐受或适应较广泛的环境梯度。
2)青藏高原湖泊沉积物细菌物种丰富度和均匀度均随盐度的增加呈显著下降趋势。盐度与细菌群落α多样性指数之间的负相关可能与部分已有生态学原理一致,即预计更极端的环境微生物群落多样性更低。这可能是由于盐度的增加影响了细胞的生理特性,增强了其渗透潜力,降低了微生物的活性和生物量,进而显著影响细菌群落α多样性。
3)湖泊沉积物细菌丰富度、均匀度和群落组成变化均受到盐度直接或间接的影响,并且受到营养盐、地理位置和气候因素的直接影响。盐度一方面直接影响沉积物细菌群落,另一方面通过影响溶解氧、TN和营养盐而间接影响细菌群落。除了盐度,营养盐、地理位置和气候因素在湖泊沉积物细菌群落中也起着重要作用。
5 附录
附表Ⅰ见电子版(DOI: 10.18307/2025.0137)。

